技术原理 / Technical Principles
创新性基因编辑技术—锐湃尔™
(REPERE ™)
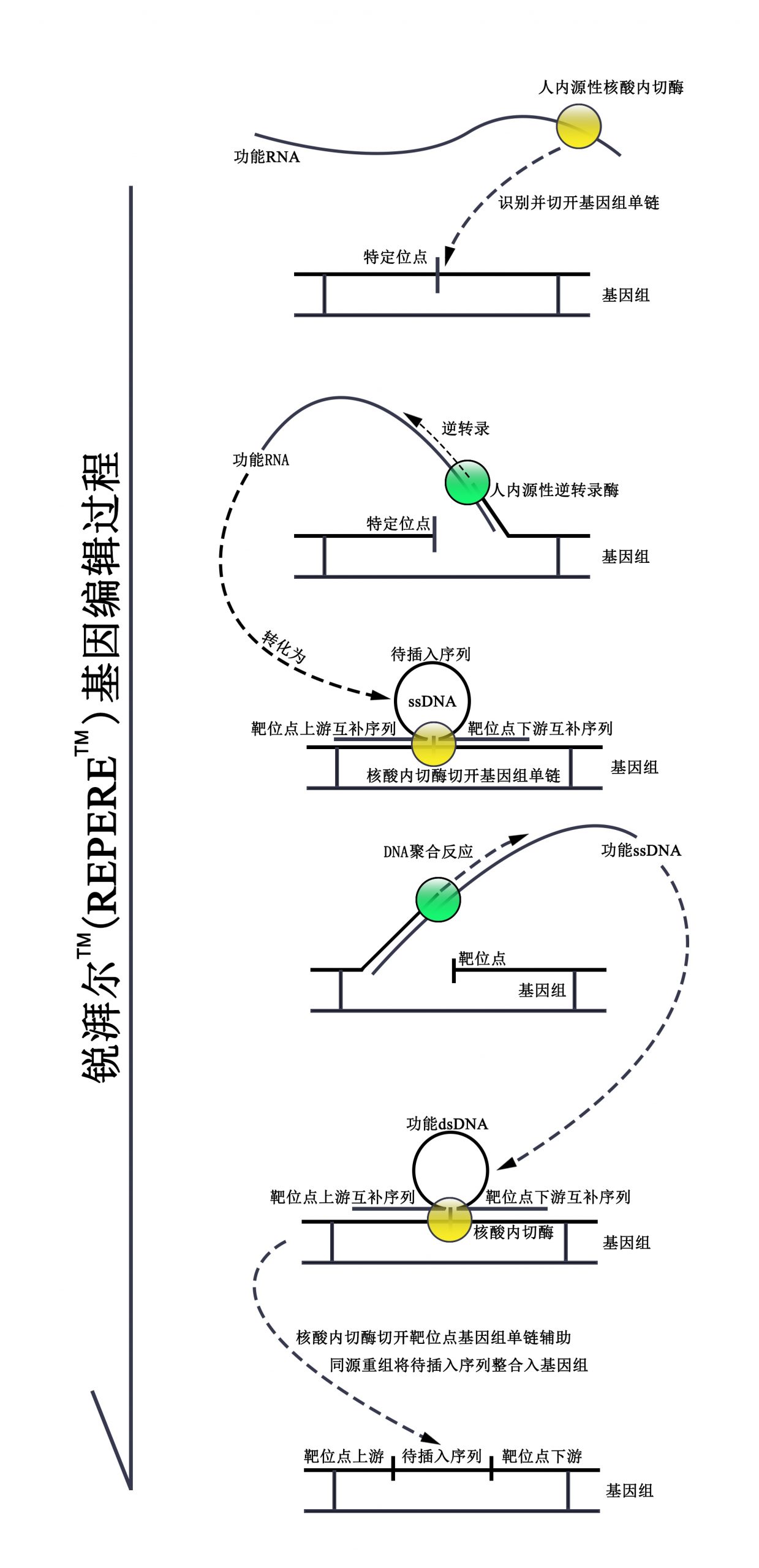
锐湃尔™应用人类基因组中的重复元件(Alu元件及LINE-1转座子)、人源核酸内切酶以及逆转录酶ORF2p,整个技术完全为人源,锐湃尔™可识别基因组上待插入序列上游的长序列片段并剪切基因组靶位点的DNA单链,同时将含有待插入序列的RNA转化为双链DNA,最终经同源重组将待插入序列插入至基因组中。该技术由于仅在待插入位点处形成双链DNA、识别序列长以及完全为人源等特点,使得其具有高靶向性、低脱靶率及极低抗原性等优势,并可以较高效率向基因组插入长片段序列。同时,该技术所采用的核酸内切酶仅切开基因组单链,此后通过同源重组进行序列插入,极大的避免了使基因组发生双链断裂的风险,因此具有较高的安全性。
锐湃尔TM技术基因编辑过程简述
含有待插入序列、靶位点上游序列及靶位点下游序列信息的RNA被由与其相连的重复元件及其所形成的特定二级结构所招募的内切酶及逆转录酶ORF2p逆转录转化为ssDNA。此后该含有待插入序列、靶位点上游序列及靶位点下游序列信息的ssDNA以序列特异性结合于基因组上靶位点处,连于ssDNA的3’端的ORF2p在ssDNA上靶位点上游序列与基因组上靶位点上游序列准确匹配后方可滑动至靶位点(该机制使得锐湃尔TM技术具有高靶向性的特点),在ssDNA在基因组靶位点处形成特定“Ω”二级结构的条件下,ORF2p切开基因组靶位点并以基因组单链为引物合成ssDNA的另一条链,转变为dsDNA。正因仅在基因组靶位点处方可形成具有同源重组潜能的dsDNA,且不产生双链断裂,锐湃尔TM技术才更为安全。此后在ORF2p的辅助下,dsDNA上的待插入序列可于细胞分裂时借助同源重组被插入至靶位点中,进而达到基因编辑的目的。锐湃尔TM技术所用序列和蛋白均取自人体,为完全人源的基因编辑技术,无需担心免疫反应;且锐湃尔TM技术基于人体固有的基因变化机制“CNV延伸机制”,安全性更有保障。
锐湃尔™可通过DNA、RNA或RNP多种形式介导基因编辑,具有较高的临床应用潜能及临床价值。

锐湃尔™研发经历
2021年1月22日
锐湃尔™技术申请发明专利,发明创造名称:基因转录框架、载体系统、基因组序列编辑方法及应用;申请号: 202110089068.1
2021年2月11日
锐湃尔™技术的基本理论“CNV延伸机制”发表于“frontiers in Cell and Developmental Biology”(目前IF:6.684)
.
CNV延伸理论合理的解释了基因组上广泛存在的内含子、短散在核元件和长散在核元件的存在意义;
同时对基因组上一系列机制尚不明朗的变化现象如胚胎和肿瘤中与相应基因表达相关的拷贝数变化、亨廷顿舞蹈症及脆性X综合症中与相应基因表达相关的三联核苷酸重复增加及仅在具有增殖潜能的免疫细胞中出现的不完整HIV基因组对高表达基因中Alu序列的偏好性插入等提出了更为合理的解释
2021年12月1日
锐湃尔™技术申请PCT,PCT申请号: PCT/CN2021/134710
技术背景简述
转座子(Alu元件及LINE-1等)及内含子占据了人类基因组上90%以上的区域,但目前绝大多数无明确作用1,2。
1.Sullivan JC, Reitzel AM, Finnerty JR. A high percentage of introns in human genes were present early in animal evolution: evidence from the basal metazoan Nematostella vectensis. Genome Inform. 2006;17(1):219-229.
2.Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome [published correction appears in Nature 2001 Aug 2;412(6846):565] [published correction appears in Nature 2001 Jun 7;411(6838):720. Szustakowki, J [corrected to Szustakowski, J]]. Nature. 2001;409(6822):860-921. doi:10.1038/35057062
LINE-1和Alu元件均为人类基因组中目前仍具有活性的转座子1,2。LINE-1可转录产生其相应RNA,表达ORF2p蛋白以助其RNA逆转录并插入(转座)回基因组1,2;Alu元件则亦可由其转录的RNA逆转录并插入(转座)至基因组,但Alu元件转座需LINE-1所表达的ORF2p辅助1,2。
1.Streva VA, Faber ZJ, Deininger PL. LINE-1 and Alu retrotransposition exhibit clonal variation. Mob DNA. 2013;4(1):16. Published 2013 Jun 3. doi:10.1186/1759-8753-4-16
2.de Andrade A, Wang M, Bonaldo MF, Xie H, Soares MB. Genetic and epigenetic variations contributed by Alu retrotransposition. BMC Genomics. 2011;12:617. Published 2011 Dec 20. doi:10.1186/1471-2164-12-617
拷贝数变异(copy number variation,CNV)在首次人类基因组测序中被严重低估1,直至近年才被逐步发掘重视。至少有数千个基因具有拷贝数变异,各基因的拷贝数变异占据了人类基因组的大块区域2-5。
1.Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome [published correction appears in Nature 2001 Aug 2;412(6846):565] [published correction appears in Nature 2001 Jun 7;411(6838):720. Szustakowki, J [corrected to Szustakowski, J]]. Nature. 2001;409(6822):860-921. doi:10.1038/35057062
2.Iafrate AJ, Feuk L, Rivera MN, et al. Detection of large-scale variation in the human genome. Nat Genet.2004;36(9):949-951. doi:10.1038/ng1416
3.Sebat J, Lakshmi B, Troge J, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305(5683):525-528. doi:10.1126/science.1098918
4.Redon R, Ishikawa S, Fitch KR, et al. Global variation in copy number in the human genome. Nature. 2006;444(7118):444-454. doi:10.1038/nature05329
5.Korbel JO, Urban AE, Affourtit JP, et al. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007;318(5849):420-426. doi:10.1126/science.1149504
在肿瘤细胞中以及发育的胚胎中,可见有规律的、特异的基因组上拷贝数变异改变,目前这些改变尚无明确且合理的机制进行解释1-8。
- Vanneste E, Voet T, Le Caignec C, et al. Chromosome instability is common in human cleavage-stage embryos. Nat Med. 2009;15(5):577-583. doi:10.1038/nm.1924
- Zhu P, Guo H, Ren Y, et al. Single-cell DNA methylome sequencing of human preimplantation embryos. Nat Genet. 2018;50(1):12-19. doi:10.1038/s41588-017-0007-6
- Claycomb JM, Benasutti M, Bosco G, Fenger DD, Orr-Weaver TL. Gene amplification as a developmental strategy: isolation of two developmental amplicons in Drosophila. Dev Cell. 2004;6(1):145-155. doi:10.1016/s1534-5807(03)00398-8
- Fischer U, Keller A, Voss M, Backes C, Welter C, Meese E. Genome-wide gene amplification during differentiation of neural progenitor cells in vitro. PLoS One. 2012;7(5):e37422. doi:10.1371/journal.pone.0037422
- Meinhardt G, Kaltenberger S, Fiala C, Knöfler M, Pollheimer J. ERBB2 gene amplification increases during the transition of proximal EGFR(+) to distal HLA-G(+) first trimester cell column trophoblasts. Placenta. 2015;36(8):803-808. doi:10.1016/j.placenta.2015.05.017
- Hannibal RL, Baker JC. Selective Amplification of the Genome Surrounding Key Placental Genes in Trophoblast Giant Cells. Curr Biol. 2016;26(2):230-236. doi:10.1016/j.cub.2015.11.060
- Shao X, Lv N, Liao J, et al. Copy number variation is highly correlated with differential gene expression: a pan-cancer study. BMC Med Genet. 2019;20(1):175. doi:10.1186/s12881-019-0909-5
- Henrichsen CN, Vinckenbosch N, Zöllner S, et al. Segmental copy number variation shapes tissue transcriptomes. Nat Genet. 2009;41(4):424-429. doi:10.1038/ng.345
随着胚胎干细胞的有丝分裂可见基因组上各基因的拷贝数变化1。
1.Liang Q, Conte N, Skarnes WC, Bradley A. Extensive genomic copy number variation in embryonic stem cells. Proc Natl Acad Sci U S A. 2008;105(45):17453-17456. doi:10.1073/pnas.0805638105
在肿瘤细胞、干细胞以及胚胎中,亦可见转座子如LINE-1及SINE的去甲基化以及高表达1-4。
1.Martin SL. The ORF1 protein encoded by LINE-1: structure and function during L1 retrotransposition. J Biomed Biotechnol. 2006;2006(1):45621. doi:10.1155/JBB/2006/45621
2.Adeniyi-Jones S, Zasloff M. Transcription, processing and nuclear transport of a B1 Alu RNA species complementary to an intron of the murine alpha-fetoprotein gene. Nature. 1985;317(6032):81-84. doi:10.1038/317081a0
3.Kerachian MA, Kerachian M. Long interspersed nucleotide element-1 (LINE-1) methylation in colorectal cancer. Clin Chim Acta. 2019;488:209-214. doi:10.1016/j.cca.2018.11.018
4.Whongsiri P, Goering W, Lautwein T, et al. Many Different LINE-1 Retroelements Are Activated in Bladder Cancer. Int J Mol Sci. 2020;21(24):9433. Published 2020 Dec 11. doi:10.3390/ijms21249433
对LINE-1的表达进行抑制后胚胎发育停滞1。
1.Percharde M, Lin CJ, Yin Y, et al. A LINE1-Nucleolin Partnership Regulates Early Development and ESC Identity. Cell. 2018;174(2):391-405.e19. doi:10.1016/j.cell.2018.05.043
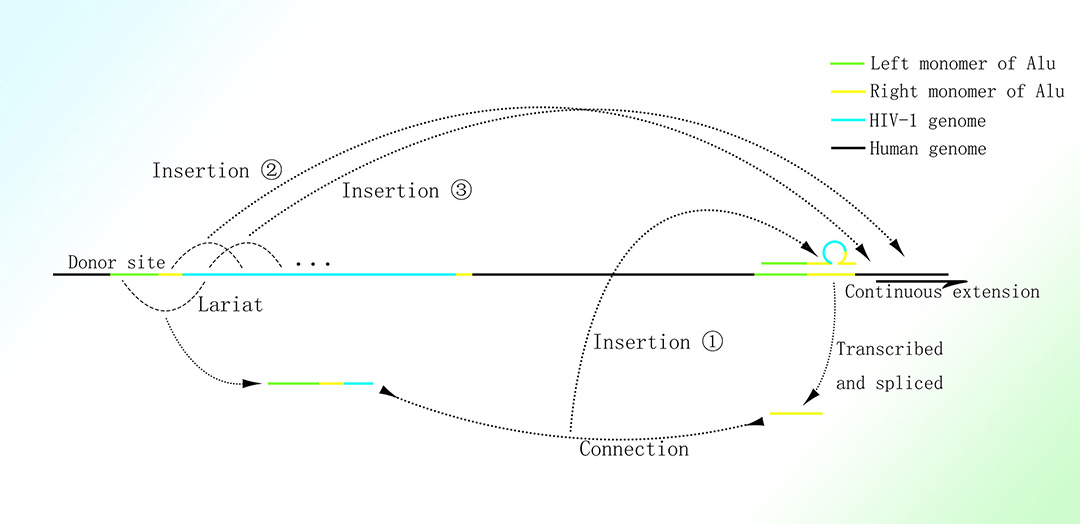
HIV基因组偏好于插入至人类基因组中的Alu元件,尤其是高转录水平基因上的Alu元件;此种克隆性扩增的插入仅发生于少数具有持续增殖能力的细胞;绝大多数被插入的HIV基因组仅为其完整基因组的一部分1-3。
1.Cohn LB, Silva IT, Oliveira TY, et al. HIV-1 integration landscape during latent and active infection. Cell. 2015;160(3):420-432. doi:10.1016/j.cell.2015.01.020
2.Maldarelli F, Wu X, Su L, et al. HIV latency. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science. 2014;345(6193):179-183. doi:10.1126/science.1254194
3.Wagner TA, McLaughlin S, Garg K, et al. HIV latency. Proliferation of cells with HIV integrated into cancer genes contributes to persistent infection. Science. 2014;345(6196):570-573. doi:10.1126/science.1256304
推定的HIV基因组在人体中的克隆性扩增机制
图片取自A Mechanism Leading to Changes in Copy Number Variations Affected by Transcriptional Level Might Be Involved in Evolution, Embryonic Development, Senescence, and Oncogenesis Mediated by Retrotransposons.
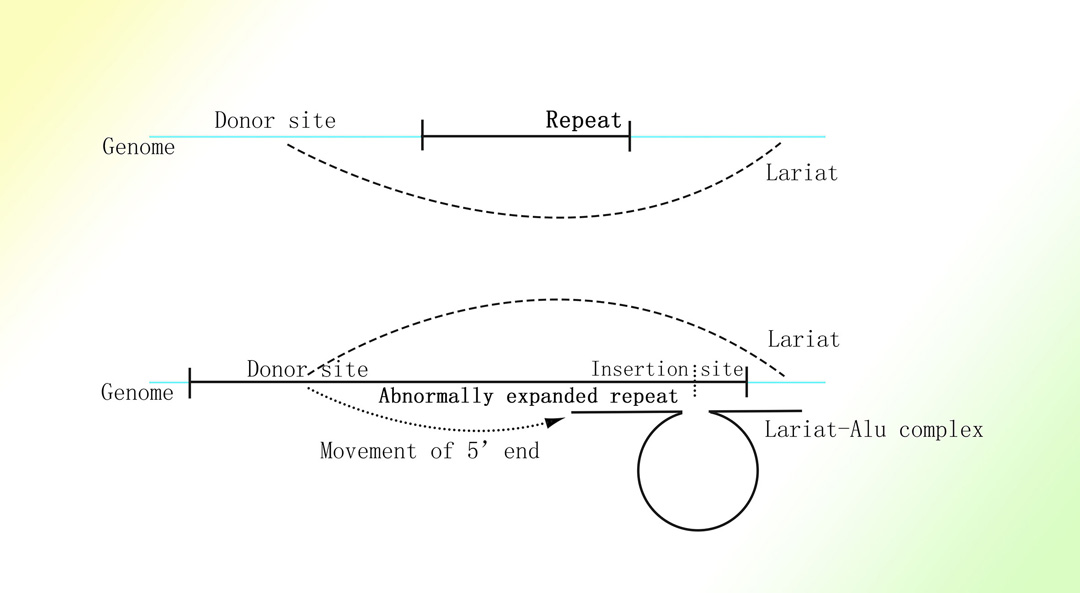
亨廷顿舞蹈症和脆性X综合症的致病基因中的重复三联核苷酸序列的扩增与相应基因的转录水平呈正相关1-4。
1.Cohn LB, Silva IT, Oliveira TY, et al. HIV-1 integration landscape during latent and active infection. Cell. 2015;160(3):420-432. doi:10.1016/j.cell.2015.01.020
2.Maldarelli F, Wu X, Su L, et al. HIV latency. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science. 2014;345(6193):179-183. doi:10.1126/science.1254194
3.Wagner TA, McLaughlin S, Garg K, et al. HIV latency. Proliferation of cells with HIV integrated into cancer genes contributes to persistent infection. Science. 2014;345(6196):570-573. doi:10.1126/science.1256304
推定的亨廷顿舞蹈症和脆性X综合症的致病基因中的重复三联核苷酸序列的扩增机制
图片取自A Mechanism Leading to Changes in Copy Number Variations Affected by Transcriptional Level Might Be Involved in Evolution, Embryonic Development, Senescence, and Oncogenesis Mediated by Retrotransposons.
内含子产生的circRNA可由内含子经剪接形成的套索结构生成1。
1.Zhang Y, Zhang XO, Chen T, et al. Circular intronic long noncoding RNAs. Mol Cell. 2013;51(6):792-806. doi:10.1016/j.molcel.2013.08.017
内含子剪切的短片段理论:将具有多聚A序列的线性RNA去除后,circRNA所检测的以剪切位点为起始或终止的circRNA(说明与内含子剪接有关)长度绝大多数<400bp。经剪接机制产生的套索结构具有3'端残端1。
1.Panda AC, De S, Grammatikakis I, et al. High-purity circular RNA isolation method (RPAD) reveals vast collection of intronic circRNAs. Nucleic Acids Res. 2017;45(12):e116. doi:10.1093/nar/gkx297
内含子剪切所形成的套索结构半衰期极短,难以实际检测1,2。目前绝大多数认知的内含子形成的套索结构均为电脑预测或推测,真正于体内验证过的套索结构不超过100个3。
1.Burnette JM, Miyamoto-Sato E, Schaub MA, Conklin J, Lopez AJ. Subdivision of large introns in Drosophila by recursive splicing at nonexonic elements. Genetics. 2005;170(2):661-674. doi:10.1534/genetics.104.039701
2.Kelly S, Georgomanolis T, Zirkel A, et al. Splicing of many human genes involves sites embedded within introns. Nucleic Acids Res. 2015;43(9):4721-4732. doi:10.1093/nar/gkv386
3.Taggart AJ, DeSimone AM, Shih JS, Filloux ME, Fairbrother WG. Large-scale mapping of branchpoints in human pre-mRNA transcripts in vivo. Nat Struct Mol Biol. 2012;19(7):719-721. Published 2012 Jun 17. doi:10.1038/nsmb.2327
选择性剪切存在于人类基因组中超90%的基因中1-2。
1.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing [published correction appears in Nat Genet. 2009 Jun;41(6):762]. Nat Genet. 2008;40(12):1413-1415. doi:10.1038/ng.259
2.Wang ET, Sandberg R, Luo S, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456(7221):470-476. doi:10.1038/nature07509
去除参与内含子剪切的剪切因子ASF/SF2后,真核细胞基因组出现明显不稳定并出现染色体外的高分子长DNA片段1。
1.Li X, Manley JL. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell. 2005;122(3):365-378. doi:10.1016/j.cell.2005.06.008
有规律的短RNA转化为双链DNA:人类细胞中普遍存在一种绝大多数长度<400bp的双链或单链DNA,其产生与其相应基因序列的转录及剪切高度相关1,2。此外,很大一部分的该短序列DNA含有短散在核元件如Alu元件序列或Alu元件富含的CpG岛2。
1.Shibata Y, Kumar P, Layer R, et al. Extrachromosomal microDNAs and chromosomal microdeletions in normal tissues [published correction appears in Science. 2012 Jun 22;336(6088):1506]. Science. 2012;336(6077):82-86. doi:10.1126/science.1213307
2.Dillon LW, Kumar P, Shibata Y, et al. Production of Extrachromosomal MicroDNAs Is Linked to Mismatch Repair Pathways and Transcriptional Activity. Cell Rep. 2015;11(11):1749-1759. doi:10.1016/j.celrep.2015.05.020
精子中独立于基因组的DNA片段被称做精子DNA片段(sperm DNA fragmentation ,SDF),该片段普遍存在于正常的精子中1。
1.Evenson DP, Darzynkiewicz Z, Melamed MR. Relation of mammalian sperm chromatin heterogeneity to fertility. Science. 1980;210(4474):1131-1133. doi:10.1126/science.7444440
Alu元件转录产生的RNA在细胞核内于其序列上的固定位点被剪切为5'部分和3'部分,5'部分半衰期较长而无明显功能,3'部分则迅速消失难以检测,提示其另有功能。3'部分留有5'端残端1,2。
1.Maraia RJ, Driscoll CT, Bilyeu T, Hsu K, Darlington GJ. Multiple dispersed loci produce small cytoplasmic Alu RNA. Mol Cell Biol. 1993;13(7):4233-4241. doi:10.1128/mcb.13.7.4233-4241.1993
2.Chu WM, Liu WM, Schmid CW. RNA polymerase III promoter and terminator elements affect Alu RNA expression. Nucleic Acids Res. 1995;23(10):1750-1757. doi:10.1093/nar/23.10.1750
Alu元件转录产生的RNA于核内剪切产生的3'部分可结合人源的内切酶及逆转录酶ORF2p,ORF2p可切开基因组单链,引发逆转录并介导转座;此前尚未有理论给出ORF2p将Alu-RNA第二条链插入至基因组的合理解释1,2。
1.Mathias SL, Scott AF, Kazazian HH Jr, Boeke JD, Gabriel A. Reverse transcriptase encoded by a human transposable element. Science. 1991;254(5039):1808-1810. doi:10.1126/science.1722352
2.Wallace N, Wagstaff BJ, Deininger PL, Roy-Engel AM. LINE-1 ORF1 protein enhances Alu SINE retrotransposition. Gene. 2008;419(1-2):1-6. doi:10.1016/j.gene.2008.04.007
Alu-RNA结合ORF2p于基因组上所作用的剪切序列绝大多数为5-TTTT/AA-3(“/”代表剪切位点),基因组上剪切序列于Alu-RNA上所对应序列的二级结构呈“Ω”型,“Ω”的缺口正对剪切位点1-3。
1.Mathias SL, Scott AF, Kazazian HH Jr, Boeke JD, Gabriel A. Reverse transcriptase encoded by a human transposable element. Science. 1991;254(5039):1808-1810. doi:10.1126/science.1722352
2.Wallace N, Wagstaff BJ, Deininger PL, Roy-Engel AM. LINE-1 ORF1 protein enhances Alu SINE retrotransposition. Gene. 2008;419(1-2):1-6. doi:10.1016/j.gene.2008.04.007
3.Häsler J, Strub K. Alu elements as regulators of gene expression [published correction appears in Nucleic Acids Res. 2007;35(4):1389]. Nucleic Acids Res. 2006;34(19):5491-5497. doi:10.1093/nar/gkl706
在人体中,转座子如LINE-1可编码逆转录酶ORF2p,对RNA进行逆转录。在LINE-1的帮助下,任何mRNA均可被逆转录并被插入(转座)至基因组中1,2。
1.Esnault C, Maestre J, Heidmann T. Human LINE retrotransposons generate processed pseudogenes. Nat Genet. 2000;24(4):363-367. doi:10.1038/74184
2.Dewannieux M, Esnault C, Heidmann T. LINE-mediated retrotransposition of marked Alu sequences. Nat Genet. 2003;35(1):41-48. doi:10.1038/ng1223
转录相关的同源重组普遍存在于哺乳动物体内,并可引起基因组改变1。
1.Gottipati P, Cassel TN, Savolainen L, Helleday T. Transcription-associated recombination is dependent on replication in Mammalian cells. Mol Cell Biol. 2008;28(1):154-164. doi:10.1128/MCB.00816-07
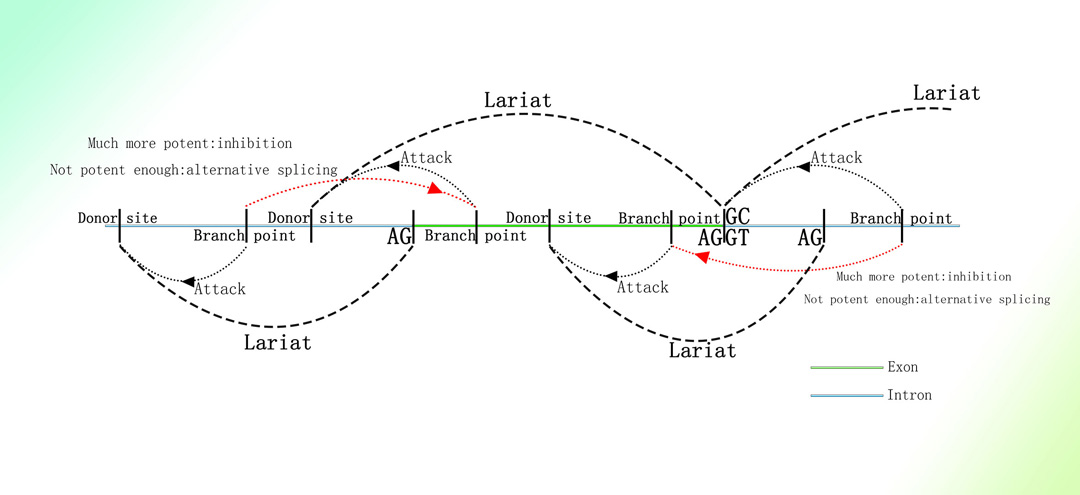
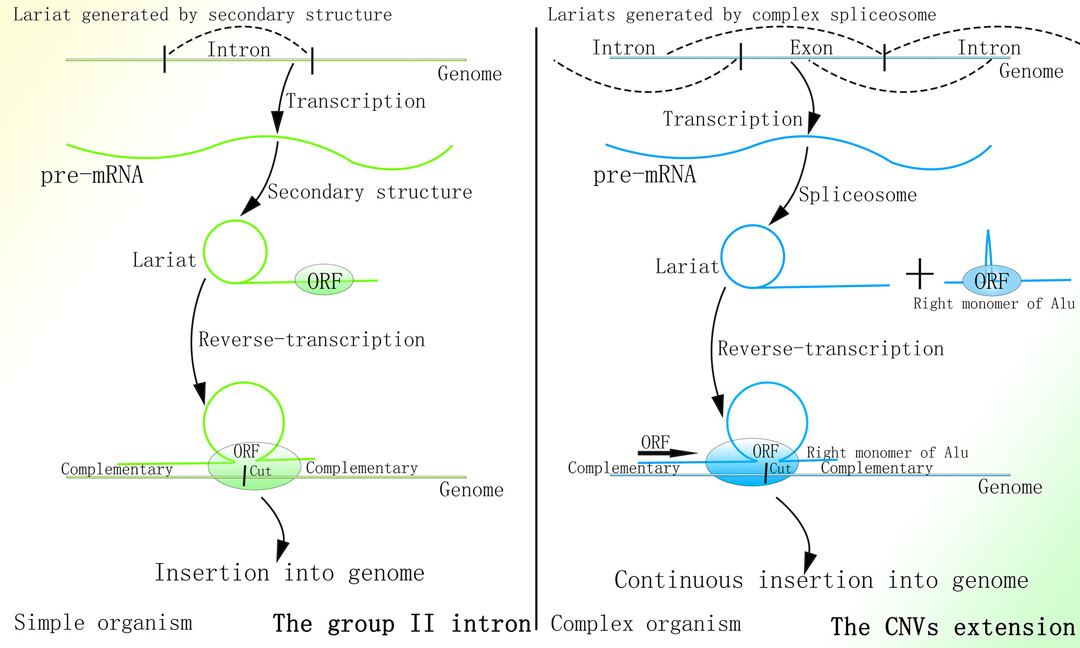
细菌中的II型内含子的转座功能依赖于其套索结构+内切酶+逆转录酶(目前Targetron基因编辑技术便基于此)1-4。
1.Guo H, Karberg M, Long M, Jones JP 3rd, Sullenger B, Lambowitz AM. Group II introns designed to insert into therapeutically relevant DNA target sites in human cells. Science. 2000;289(5478):452-457. doi:10.1126/science.289.5478.452
2.Guo H, Zimmerly S, Perlman PS, Lambowitz AM. Group II intron endonucleases use both RNA and protein subunits for recognition of specific sequences in double-stranded DNA. EMBO J. 1997;16(22):6835-6848. doi:10.1093/emboj/16.22.6835
3.Jiménez-Zurdo JI, García-Rodríguez FM, Barrientos-Durán A, Toro N. DNA target site requirements for homing in vivo of a bacterial group II intron encoding a protein lacking the DNA endonuclease domain. J Mol Biol. 2003;326(2):413-423. doi:10.1016/s0022-2836(02)01380-3
4.Singh NN, Lambowitz AM. Interaction of a group II intron ribonucleoprotein endonuclease with its DNA target site investigated by DNA footprinting and modification interference. J Mol Biol. 2001;309(2):361-386. doi:10.1006/jmbi.2001.4658
II型内含子可首先由RNA转化为dsDNA后基于同源重组进行转座归巢1。
1.Cousineau B, Lawrence S, Smith D, Belfort M. Retrotransposition of a bacterial group II intron [published correction appears in Nature 2001 Nov 1;414(6859):84]. Nature. 2000;404(6781):1018-1021. doi:10.1038/35010029
人体内已有RNA经内源性逆转录酶逆转录为双链DNA,并随细胞分裂通过同源重组定向且特异的插入至基因组的先例,即端粒酶延长端粒的相关机制1,2。此外,在某些缺失端粒酶的物种中,转座子也可替代端粒酶进行端粒的延长3,4。
1.Shampay J, Blackburn EH. Generation of telomere-length heterogeneity in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1988;85(2):534-538. doi:10.1073/pnas.85.2.534
2.Sobinoff AP, Pickett HA. Mechanisms that drive telomere maintenance and recombination in human cancers. Curr Opin Genet Dev. 2020;60:25-30. doi:10.1016/j.gde.2020.02.006
3.Danilevskaya ON, Arkhipova IR, Traverse KL, Pardue ML. Promoting in tandem: the promoter for telomere transposon HeT-A and implications for the evolution of retroviral LTRs. Cell. 1997;88(5):647-655. doi:10.1016/s0092-8674(00)81907-8
4.Villasante A, de Pablos B, Méndez-Lago M, Abad JP. Telomere maintenance in Drosophila: rapid transposon evolution at chromosome ends. Cell Cycle. 2008;7(14):2134-2138. doi:10.4161/cc.7.14.6275
锐湃尔™所基于的CNV延伸机制与端粒酶延伸端粒的相关机制均是通过细胞内的内源性逆转录酶逆转录核内转录的RNA为双链DNA并定向插入至基因组的特定区域都是通过细胞分裂来限制由其相应机制介导的基因组改变。
锐湃尔™技术所基于的CNV延伸机制与II型内含子的转座机制则均是先使内含子RNA通过特殊的二级结构形成套索结构,此后通过结合于套索结构的内切酶切开基因组同时将RNA逆转录为双链DNA,最终通过双链DNA与基因组结合形成“Ω”型结构并通过同源重组插入至基因组。
II型内含子与锐湃尔™技术比较
图片取自A Mechanism Leading to Changes in Copy Number Variations Affected by Transcriptional Level Might Be Involved in Evolution, Embryonic Development, Senescence, and Oncogenesis Mediated by Retrotransposons.